We just sequenced another 122 Cannabis microbiomes in collaboration with DigiPath labs in NV to try to resolve differences between 3M and qPCR methods. We focused on samples that were discordant where 3M finds something but qPCR misses it or vice versa.
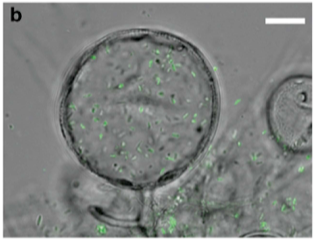
In this case we went and sequenced individual colonies on 3M Total Yeast and Mold films and found both Bacteria and Fungi present in the colonies. The colonies would PCR very efficiently with our 16S and 18S primers suggesting the failure to detect the microbes seen on films was not a result of primer design but likely a result of sampling bias. This will make more sense once you understand the lifecycle of this Bacteria.
In almost all circumstances we find a bacteria known as Ralstonia.
Ralstonia is a plant and human pathogen and it can infect other Fungi like Aspergillus, Penicillium and Fusarium. Once it infects the fungi it causes the fungi to make Chlamydospores. This is a multicellular sporulation stage that clumps making sampling for testing very challenging.
Ralstonia lung infections do happen in cancer wards, ventilators, Cystic Fibrosis patients and immunocompromised patients.
This presents an ugly challenge for 3M petri dishes. How can one assume 10,000 CFU/g of Yeast and Mold is safe if any of those fungi can have thousands of virulent endofungal bacteria? Would this bacterial risk be picked up with TAC plating if the bacteria is endogenous to a Fungi? The capacity to properly quantify CFU/g assumes a single cell created a colony. This is not true with filamentous fungi or clumped chlamydospores.
Another interesting example of dangerous endofungal bacteria is Rhizopus and Rhizoxin production. This toxin is made by an endofungal bacteria.
Anyone have experience with Ralstonia?
https://app.onecodex.com/analysis/public/f489bf6a036948dc
https://www.ncbi.nlm.nih.gov/m/pubmed/26943626/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5297155/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4086841/
https://www.sciencedirect.com/science/article/pii/S0195670105003713