My fascination with this topic began after meeting artist Mark Curtis in 2017. Driven by his interest in nature and the depiction of space, Curtis began an investigation into structure of DNA in 1995. The resulting artwork was later used for a Royal Mail 'Millennium Collection' stamp, commemorating the discovery of DNA.
The following is taken from New Scientist, 16 May 1998 - Deconstructing DNA:
After many attempts, he found he couldn't make sense of the two and three-dimensional representations he found in textbooks. They seemed to contradict one another. Frustration finally forced him back to what he refers to as "geometrical first principles", and in the manner of Renaissance perspectival artists such as Paolo Uccello, he began by making careful scale drawings of DNA.
DNA has ten base pairs to each turn of the helix, so, using these centuries-old methods, Curtis began with a decagon-a 10-sided figure. He then placed a pentagon for each base around it. The pentagon is the only regular shape that can fit round a decagon in this way without leaving any space. And according to Curtis, this figure of 10 pentagons oriented about a decagon is the only geometrical configuration that enables one to create a helix with the known dimensions of DNA. Add some thickness to each of the pentagons, and the 10 pentagons telescope out to form a helix. To build a structurally sound double helix, Curtis reasoned that he would need not just one pentagon, but two joined together. With this insight he was finally able to produce the drawings and paintings that he had originally planned.
This kind of reasoning may seem obscure by modern standards. But for Curtis, it established that a double helix of 10 turns had to be made of twinned pentagons. At this point in his work, he was still thinking in terms of the traditional Watson and Crick structure. But he faced a contradiction: in the standard Watson and Crick chemistry, the base pairs join through hexagonal regions in their structures. Then came Curtis's moment of truth: "I sat down on the sofa one night and I thought, hang on a second, the molecular structures of the bases also have pentagons in them. And here was I, with two pentagons, building a consistent helix that conformed to the dimensions of the DNA double helix." By connecting the bases differently, Curtis found that he could naturally form pairs of pentagons in each base pair. "It was placed in my lap. I wasn't trying to prove anybody wrong. I wasn't even thinking then that they've got it wrong. I was just playing, like artists play."

In Curtis's painting, ten pentagons arranged around a decagon form a helix with the known dimensions of DNA
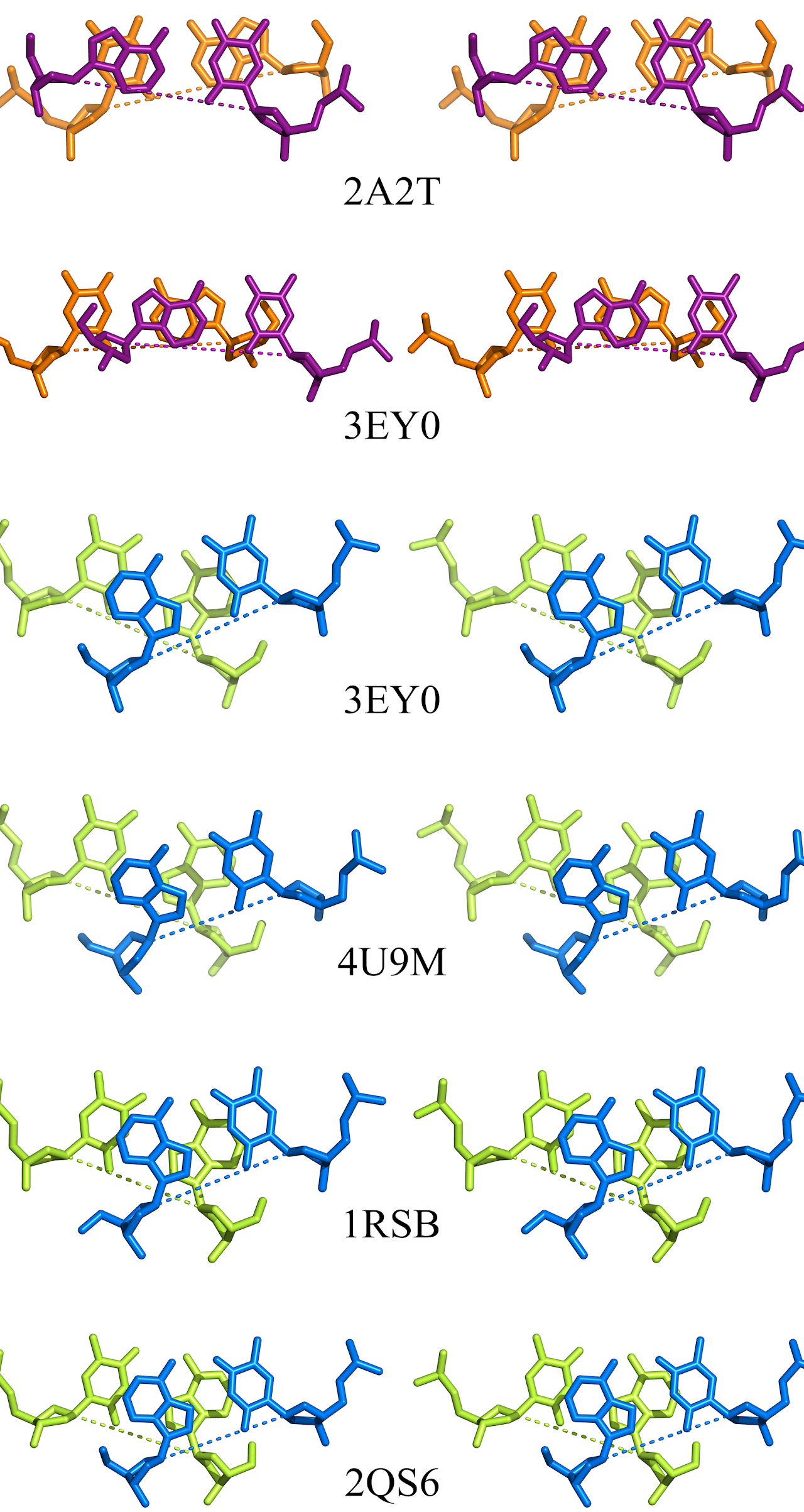
Striking similarities to Curtis' pentagonal stacking, from: Structure of the DNA Duplex d(ATTAAT)2 with Hoogsteen Hydrogen Bonds
It was later brought to Curtis' attention that he had inadvertently created base pairs discovered by Karst Hoogsteen in 1959.
There is a growing body of work on Hoogsteen base pairs. The following is from a 2019 paper - Infrared Spectroscopic Observation of a G–C+ Hoogsteen Base Pair in the DNATATA‐Box Binding Protein Complex Under Solution Conditions:
Hoogsteen DNA base pairs (bps) are an alternative base pairing to canonical Watson–Crick bps and are thought to play important biochemical roles. Hoogsteen bps have been reported in a handful of X‐ray structures of protein–DNA complexes. However, there are several examples of Hoogsteen bps in crystal structures that form Watson–Crick bps when examined under solution conditions. Furthermore, Hoogsteen bps can sometimes be difficult to resolve in DNA:protein complexes by X‐ray crystallography due to ambiguous electron density and by solution‐state NMR spectroscopy due to size limitations. Here, using infrared spectroscopy, we report the first direct solution‐state observation of a Hoogsteen (G–C+) bp in a DNA:protein complex under solution conditions with specific application to DNA‐bound TATA‐box binding protein. These results support a previous assignment of a G–C+ Hoogsteen bp in the complex, and indicate that Hoogsteen bps do indeed exist under solution conditions in DNA:protein complexes.
Through subsequent research I came across Professor Tai Te Wu. Wu sent me the document Politics Dictates Scientific Truth which gives an overview of his experience. All of Wu's publications along with correspondence between Wu and Maurice Wilkins can be found on this page.
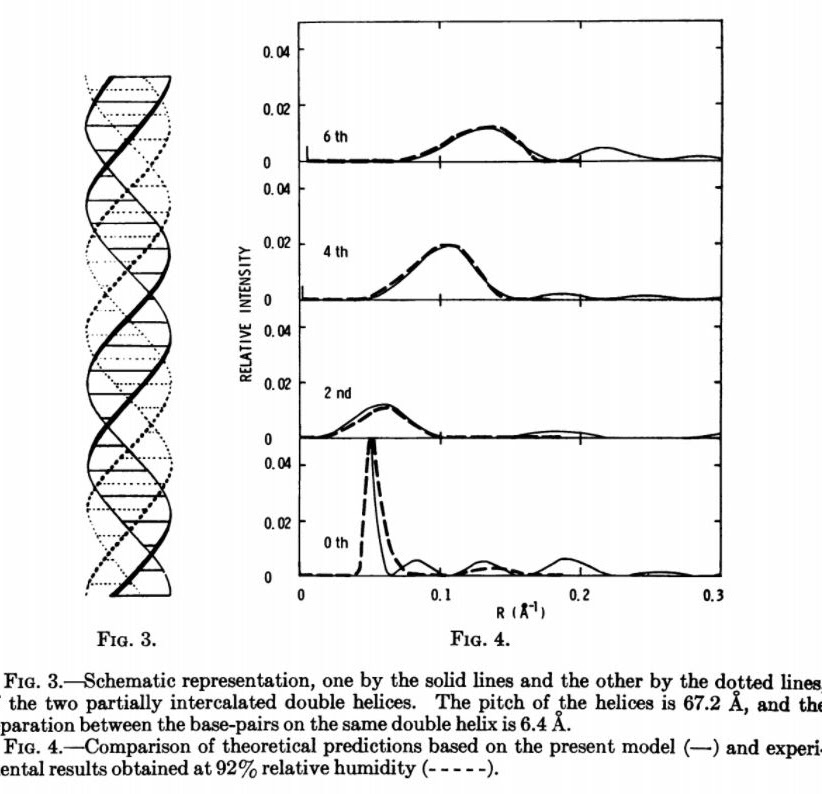
In Secondary Structures Of DNA, Wu proposes an alternative structure to explain the diffraction pattern in Photo 51:
Through systematic search, a four-stranded structure that gives predicted diffraction patterns in agreement with the experimental result at 92 per cent relative humidity has been obtained. It is schematically illustrated in Figure 3. The solid lines represent one base-paired double helix, which can be considered as a stretched-out version of the conventional double helix. Its pitch is 67.2 A, and the separation between base-pairs is 6.4 A. The pairing is of the Hoogsteen type. The other identical double helix is represented by dotted lines. It is located symmetrically opposite to the helical axis. These two double helices are held together by partial base-pair intercalation. As a result, the effective pitch is reduced to 33.6 A, and the effective separation between base-pairs is 3.2 A.
The comparison between experiment and theory is shown in Figure 4. Since the pitch of the helices is twice that of the conventional one, there should be twice the number of layer lines. But, by symmetry, the intensities on odd-layer lines vanish within the region of interest. The theoretical 0, 2nd, 4th, and 6th layer lines therefore correspond to the experimental 0, 1st, 2nd, and 3rd layer lines. The agreement is indeed remarkable. A comparison with Figure 2 clearly indicates that the predicted diffraction patterns based on the present model can only fit the experimental result obtained at 92 per cent relative humidity but not at 66 per cent.

Curtis' models adopting the intercalated structure proposed by Tai Te Wu
Dr Ken Biegeleisen developed Wu's ideas further. His papers can also be found here, along with accompanying video presentations.
Wu and Biegeleisen are mainly concerned with the net helicity and tertiary structure of DNA. Wu published the paper - "A novel intact circular dsDNA supercoil" in 1996 which has been largely ignored:
The combination of the above theoretical model and experimental results strongly suggests that there is an alternative structure of DNA which does not have the usual difficulty of unwinding, rewinding and requiring numerous covalent bond breakages and ligations during semiconservative replication.
Biegeleisen proposes a different means to test this theory in Methods for Non-Destructively Separating or Reannealing the Strands of Circular Duplex DNA Chromosomes.
I was intrigued to discover that Curtis' models are able to adopt all the forms put forward by Wu and uploaded a brief video demonstrating this.
Hi! I am a robot. I just upvoted you! I found similar content that readers might be interested in:
Source
Copying/Pasting content (full or partial texts, video links, etc.) with adding very little original content is frowned upon by the community. Repeated copy/paste posts could be considered spam. Spam is discouraged by the community and may result in the account being Blacklisted.
If you believe this comment is in error, please contact us in #appeals in Discord.