Isolation and adoption drives the emergence of beer yeasts
Brettanomyces on an agar plate. Image credit and license: Bojan Žunar / CC BY-SA (https://creativecommons.org/licenses/by-sa/4.0).
Last week, I described the vast diversity of beer styles, some of which feature only a relatively small cast of yeast strains. While we can experiment with commercially available yeast strains and mash-up existing recipes, one could also consider adopting lesser-known yeast species or strains. The key question, however, is: are there any other strains out there that can be used for brewing?
In this instalment of The Beerologist, I will describe a scientific paper by Prof. Dr Jochen Förster, Director of Yeast and Fermentation at the Carlsberg Research Laboratory and his team. (https://www.frontiersin.org/articles/10.3389/fmicb.2020.00637/full#h7).
What is the paper about?
His group aimed to understand the genetic diversity and brewing capabilities of the Brettanomyces genus, comprising species such as B. bruxellensis, B. anomalus, B. custersianus, and B. naardenensis. For this, they sequenced and obtained an extensive collection of 84 strains, isolated from various sources (e.g. wine, wood, wild, farmhouse beers, lambic beers etc) used in different countries and compared their sequences (the instructions that specify the makeup within, and the functions carried by a cell). Importantly, the team tested each strain in the lab for its ability to ferment sugars, produce desirable compounds (that give beer this wonderful aroma and taste) along with those chemicals that result in off-flavours. In doing so, this work has revealed some key insights.
Comparing Yeast genome blueprints reveals relationships between strains and their origins.
Every organism has its own book of instructions, written in 4-letter code. The same is true for yeast. When you compare yeast strains, however, you may find minor variations and sometimes extra pages of text (or missing chapters). Assuming that all strains have a common ancestor and those disagreements (mutations, deletions and insertions) occurred over time, you can use the information to discriminate between strains or help classify groups of isolates based on (dis)similarity. When the authors used genetic information from each strain to group them, they found distinct clusters of isolates (referred to as “clades”). When looking at characteristics, shared by members within each clade, the origin of strains, substrate and their use could explain differentiation. They found that in some clades, the isolation and use of yeast in brewing may have imposed selection pressures that reduced genetic diversity. They found strains used for brewing Lambic beers to be more diverse, consistent with the fact that its production relies on spontaneous fermentation and selection is not as stringent. Strains isolated from wine (where they are contaminants), were more diverse, yet distinct enough to fall into a single clade of related strains. The key question becomes, what sets those clades apart?
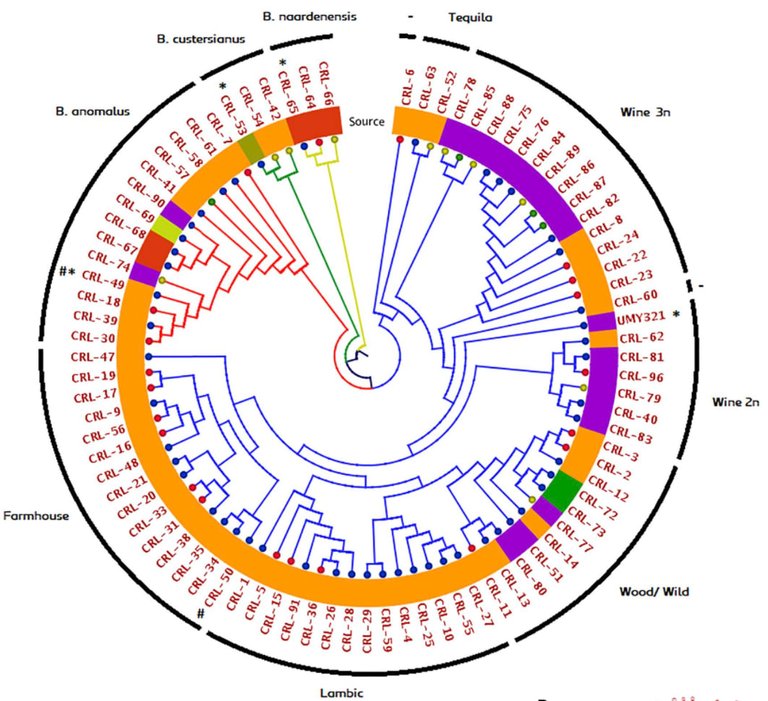
Figure 2A. reproduced from Colomer and authors (2020) showing relationships between Brettanomyces species and isolates. Clusters represent clades that have a close genetic relationship with each other. Annotation of the outer ring specifies the source of each yeast strain & group (Lambic, farmhouse beer etc). From this figure, it is clear that there are correlations between strain origin, use and species. (license).
Presence & absence of Brewing associated genes across the Brettanomyces genus.
If there is selection pressure acting on a yeast population that allows them to thrive in a brewing environment, one can expect for there to be differences in the composition or properties of genes that encode proteins that affect the process. One way of identifying such differences is to look at all the known “brewing” genes and ask: Which ones are present in the strains we know of and is there any correlation with brewing performance?
The group tried to answer this question by first identifying and describing all known brewing-associated genes in B. bruxellensis, using the UMY321 reference genome. The authors then used this sequence information to search for these genes in all other sequenced strains, using sequence similarity. These analyses revealed that some genes required for maltose uptake and breakdown are lacking in some clades, whereas in other yeast groups, genes that confer alcohol tolerance was present. These results underscore how the isolation of yeast strains from diverse sources and their adoption in beer or wine production helps shape the gene repertoire.
Importantly, the authors did not stop after completing a set of computational experiments. rather, a series of test fermentations, staining methods and growth analyses were used to tease out yeast characteristics and link them to genetic signatures identified in each given clade. In doing so, the authors established a powerful platform that allows the identification and characterisation of new yeast strains. In addition, this approach can also identify characteristics that are not desirable or require improvement. In this context, the identification of a Brettanomyces strain, lacking a key gene required for the production of off-flavour phenolics, is a case in point. Phenotypic and genome analyses revealed that the B. anomalus strain CRL-90 misses the BaPAD1 gene. BaPAD1 is responsible for the decarboxylation of ferulic acid to 4-VG, a major off-flavour compound. For this and its other characteristics (alcohol tolerance, fermentation performance and growth), this isolate emerged as a promising candidate for adoption.
Outstanding questions:
As with any important piece of work, excellent answers lead to new and interesting questions. For example, with the analyses of strains that mainly originate from Belgium, Denmark and the USA, sampling of strains across the globe may bring some nuance and new insight into Brettanomyces biology.
Similarly, with most yeast isolates originating from a brew setting, genome sequencing and comparative analyses of additional Brettanomyces strains, isolated from the natural environment will permit the identification of factors that allow adaptation to fermentative processes. What predisposes a wild yeast isolate to become a good fermenter? If there are such factors, are they in any way useful for predicting brew value?
Finally, more detailed information on wild strains, such as the location and substrate they were isolated from, would yield valuable insight into metabolic capacity. What do Brettanomyces strains consume in their natural environment and to what extent contribute (secondary) metabolites to survival in the wild? Is such information useful to inform targeted yeast capturing exercises?
As you will realise, there is a lot to learn about the biology of brewing yeasts. We are now in an exciting era where the detailed characterisation of new strains has become relatively easy. There is little doubt that new yeast species, suited for use in beer brewing, will come to the fore. We just have to go out and find them!
The Beerologist aims to inspire your journey in great beer making. Please sign up for our newsletter if you want to hear the latest in brew science! Oh, and it would be great if you could spread the word from the Beerologist far and wide!
Cheers,
Edgar, From the Beerologist
This was a very interesting read and reminded me of grapes.
There are different types of grapes, and some are better for making jams and jellies and other are better for making wines. However, the roots of most of the grapes in the world are "jelly grapes" because at one point, all of the wine grape roots were dying, and the roots were made more hearty by reinforcing them with the roots of the "jelly" grapes.
This sparked that memory because of the US yeast - was the US yeast studied yeast from Native US or was it yeast that was imported and then mixed with the land?
Hi @metzli,
That's a good question. The authors used a collection of strains, collected at various locations. The paper talks about this and even shows a figure (see below). As you can see, many isolates are from the US, but there are also a significant number from other countries in Europe (Belgium and Denmark). I think this is more a historical thing though. Brettanomyces species are found nearly everywhere. It is just a matter of where you wish to look and isolate them from.
Thank you for your comment and vote!
Best wishes,
Edgar.
Figure 1 from Colomer et al. (2020)
@tipu curate
Upvoted 👌 (Mana: 65/78) Liquid rewards.
Congratulations @huitemae! You have completed the following achievement on the Hive blockchain and have been rewarded with new badge(s) :
Your next target is to reach 900 upvotes.
You can view your badges on your board and compare yourself to others in the Ranking
If you no longer want to receive notifications, reply to this comment with the word
STOPCheck out the last post from @hivebuzz: