Hey guys! I'm back, for now, and I've decided to start my journey back into Steemit by telling you a little bit about what I'm doing in my thesis. Case in point - Molecular docking!
I'll be giving you the rundown on what this is, why it's important, and what impact it may have in the future.
Let's get started, shall we?
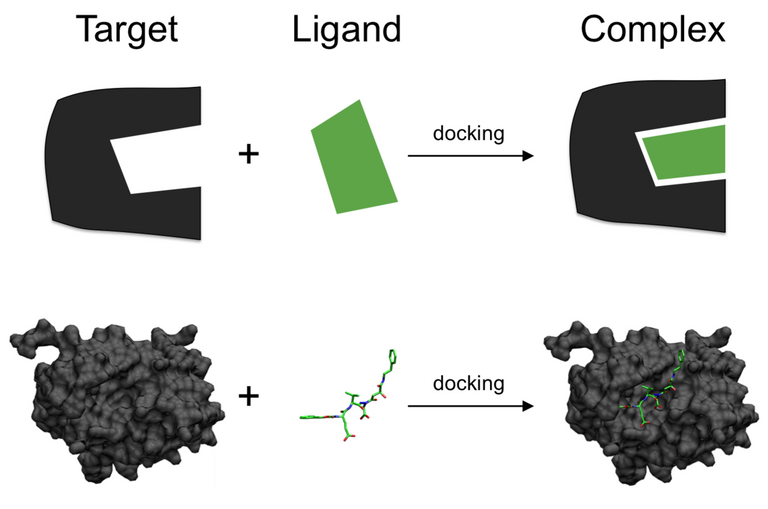
1 - Docking is a technique that predicts how molecules will connect to each other
Often, in anything involving interactions between molecules, scientists often ask themselves a very important question - "How do these two molecules connect to each other?". That might seem irrelevant to you, but it's actually of supreme importance.
You see, molecules can connect to each other in many different ways. The way they do impacts how stable the final structure will be. Therefore, by judging how different molecules connect, we can see what molecules have greater affinity between each other, and how well they would connect.
But what impact does this have in real life, you may ask?
** 2 - This technology has many applications for society**

As you may have realized, this technology holds in itself the potential to greatly enhance any research areas that involve interactions between two or more molecules.
...That's a lot of research areas actually.
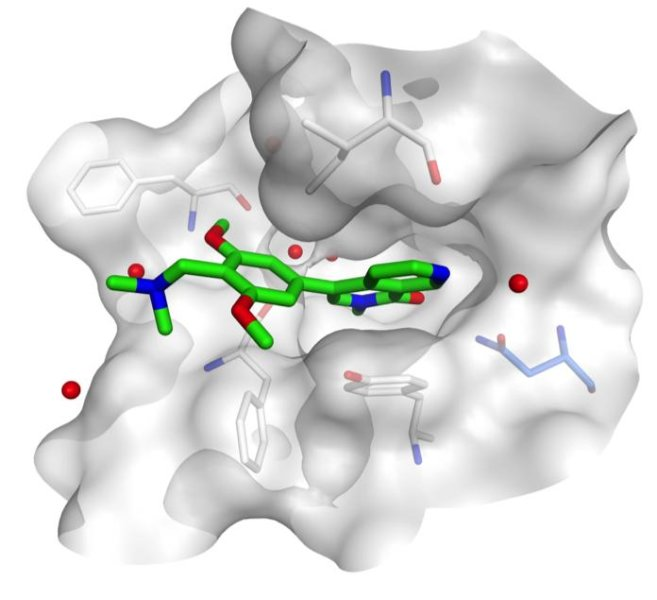
For starters, it helps design new drugs. By seeing how drugs interact with biological targets in our body, scientists can have an idea on how to change the drug's structure to maximize its' impact.

It also helps test how the drugs adhere to many different biological targets. For instance, a drug that binds to proteins will obviously have a different structure from one that binds to DNA. This process can help optimize many of the drugs existing on the market.
...Hopefully I've sold you on how useful this technique is. However, by now you may be thinking, how exactly does this technique work?
3 - Docking can be done in 2 different ways
There are different procedures you can follow when doing a docking test. Most of them fit under 2 categories - Shape complementarity and simulation.
Shape complementarity, or geometric matching, you simply isolate the main features of the two molecules. Like "molecule A has this nice groove here", or "molecule B has this nice flat area on the side". The computer takes those "descriptions", and does some quick maths to determine what is the best way those could hipothetically fit.
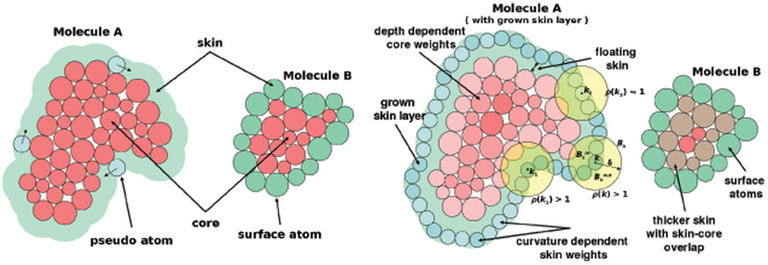
Sounds easy? Computationally, it is. Computers can go through lists of thousands of molecules in these terms, and find the best pairs, and how they pair to each other. However, the subtle nuances of a real system moving are often lost, and as such, this method isn't the most accurate.
Simulation on the other hand, takes the two molecules you give it, puts them in a box together, and just directly plays out what would happen in a real environment. From the moment the two molecules start moving towards each other, to the moment they fit in a nice little pattern, this method plays everything out in minute detail.
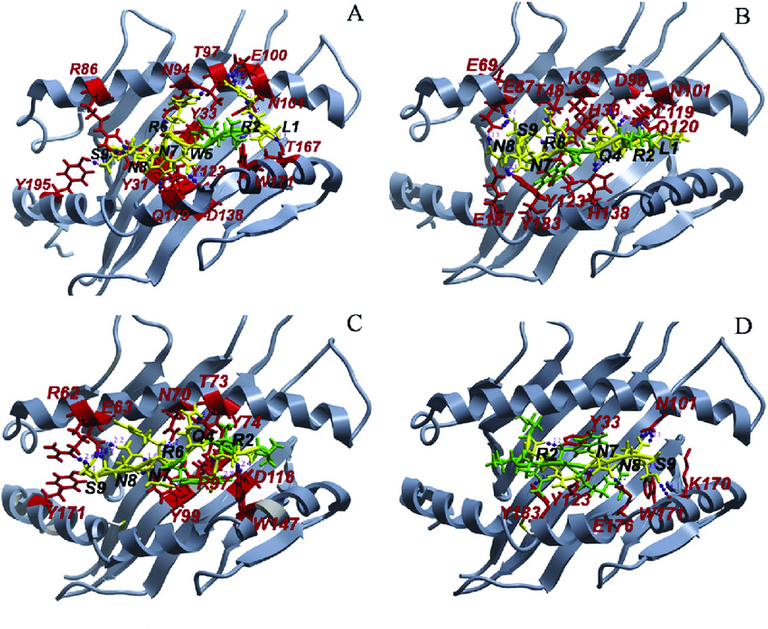
....That makes computers bust a sweat, but the results are well worth it. Certainly far more than shape complimentarity... so long as you don't mind waiting a couple of days for those results.
That said, how exactly does the computer do both of these things?
4 - Docking involves search algorithms and scoring functions
Sounded like gibberish to you? Let me explain.
A search algorithm is a nice little tool that basically tells you all the positions a molecule can be in at a given time. In both of the approaches described above, the docking program you're using will just take the data, and calculate every possible position that these two molecules could be connected in.
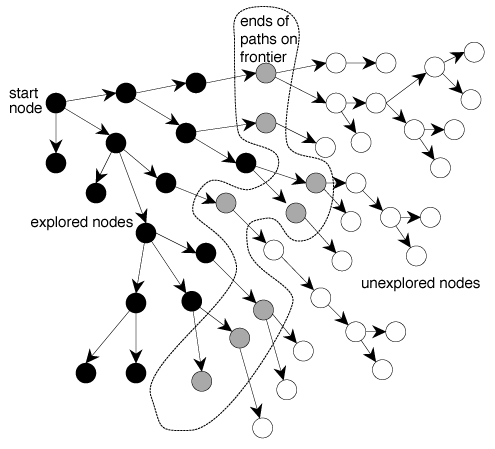
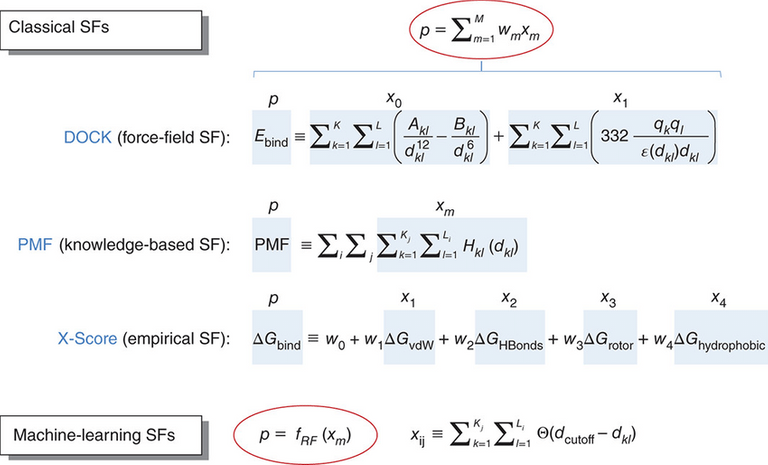
...That's pretty neat, but it doesn't tell you which positions are better, does it? That's where scoring functions come in. These functions take every single one of the positions generated by the search algorithm, and assigns a score to it. That score measures how good that connection is for both the molecules.
That score is calculated based on many factors. Free energies, rotations.... it's impossible to tell you more about that without a chemistry lecture neither of us have time for. But the gist of it is this - the better the score, the more likely that that position would occur in real life. Therefore, the program can tell what of the many possible positions it generated would happen in reality.
Convoluted? It is, a bit. Take the time to reread if you want, but that's basically it. Now, you may be thinking, how exactly do I work with this in my thesis? Good question. Which requires a detour into the world of DNA... but that's a story for next time!
That wraps this up!
Hopefully, I've given you a little bit of an inkling of what I've been doing, and sparked your interest. If I did, feel free to sound off in the comments below, I'll be very happy to answer any questions you have!
I hope I can get back to posting on a regular basis, and am very happy to be back, if only temporarily!
Till the next one guys! Steem on!