Dear Steemit Friends,
In my previous article, I talked a bit on CRISPR and I have looked for good articles here on steemit that might explain the Biology of CRISPR. It was far and not complete, I am sure there are better articles outside the ecosystem. But in an effort to bring good content here, I took up the opportunity to make a short post on CRISPR which is Part 1 of 3 part series.
The Introduction.
Before its mainstream adaptation into a genome editing technology in eukaryotes. CRISPR, an acronym for Clustered Regularly Interspaced Short Palindromic Repeats; was discovered in the genomes of bacteria and archea by the team led by Phlippe Horvath. It was described for the first time in a paper published in the journal Science on 23 March 2007. The paper titled "CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes"; As a mechanism of resistance against bacteriophages.
Bacteriophages are abundant viruses that primarily "infect" bacteria. They are ubiquitous and abundant in nature and are very important in ecological and evolutionary aspects. Since they attack bacteria; as a countermeasure, bacteria have developed their own defense mechanisms. CRISPR is one such countermeasure that was described in the article.
The discovery
The team used abacteriumm commonly used in industries; Streptococcus thermophilus as the model species. They compared closely related strains and identified that there were sites that varied a lot between species. Since the industrial species lack the resistance to bacteriophages and their wild cousins are resistant to the phage infection; They analysed these stretches of repeated sequences.
"CRISPR loci typically consist of several noncontiguous direct repeats separated by stretches of variable sequences called spacers and are oftentimes adjacent to cas genes (CRISPR-associated)"
Upon analysis, the phage resistant types had additional spacers in the CRISPR loci. To test their hypothesis that this was a sort of immune system; They challenged a strain found in the industry with different phages.
- There was addition of spacer sequence after the "infection".
- The addition was polarized to one end of the CRISPR locus.
- These sequences seemed to be obtained from the phage genome.
- These reveal that on becoming resistant to bacteriophages, CRISPR locus was modified by integration of spacers derived from phage DNA.
The authors then made classical studies of deleting the obtained spacers and introducing spacers to test the sensitivity against the resistance.
- Spacers have the ability to provide resistance de novo.
- A genetic context was necessary for the spacers to work and be effective.
- The cas genes are involved in providing the genetic context. They act as nucleases, may aid in insersion of new spacers.
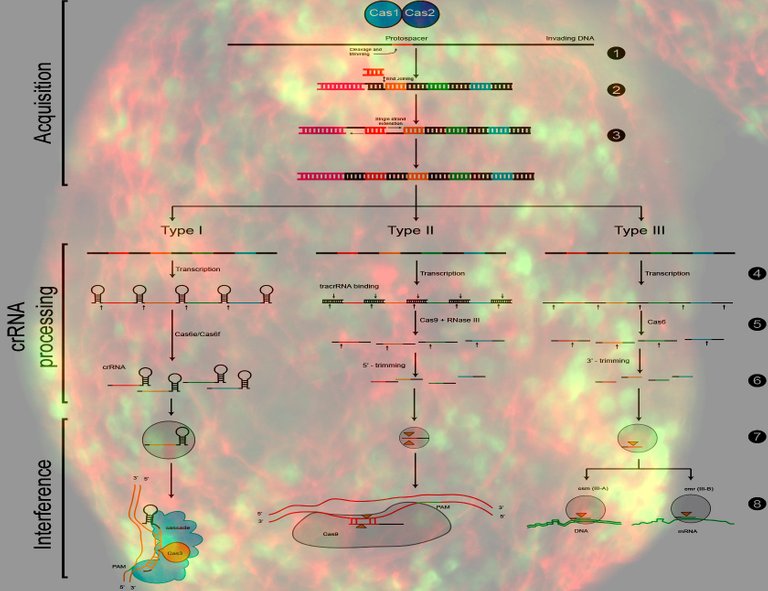
Image: By CtSkennerton (Own work), via Wikimedia Commons. Background by NIH
The stages of CRISPR immunity for each of the three major divisions.
(1) Acquisition begins by recognition of invading DNA by Cas1 and Cas2 and cleavage of a protospacer.
(2) The protospacer is ligated to the direct repeat adjacent to the leader sequence and
(3) single strand extension repairs the CRISPR and duplicates the direct repeat. The crRNA processing and interference stages occur differently in each of the three major CRISPR systems.
(4) The primary CRISPR transcript is cleaved by cas genes to produce crRNAs.
(5) In type I systems Cas6e/Cas6f cleave at the junction of ssRNA and dsRNA formed by hairpin loops in the direct repeat. Type II systems use a trans-activating (tracr) RNA to form dsRNA, which is cleaved by Cas9 and RNaseIII. Type III systems use a Cas6 homolog that does not require hairpin loops in the direct repeat for cleavage.
(6) In type II and type III systems secondary trimming is performed at either the 5’ or 3’ end to produce mature crRNAs.
(7) Mature crRNAs associate with Cas proteins to form interference complexes.
(8) In type I and type II systems, basepairing between the crRNA and the PAM causes degradation of invading DNA. Type III systems do not require a PAM for successful degradation and in type III-A systems basepairing occurs between the crRNA and mRNA rather than the DNA, targeted by type III-B systems.
The original paper can be accessed on Science Magazine.
Image Sources: All the images used are attributed to the corresponding authors. They are not mine. Please take time to check their galleries as it gives them more exposure and you will get to know their work better.

Congratulations @harshameghadri! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on any badge to view your own Board of Honor on SteemitBoard.
For more information about SteemitBoard, click here
If you no longer want to receive notifications, reply to this comment with the word
STOPHi! I am a robot. I just upvoted you! I found similar content that readers might be interested in:
https://en.wikipedia.org/wiki/CRISPR
@Originalworks
Some similarity seems to be present here:
This is an early BETA version. If you cited this source, then ignore this message! Reply if you feel this is an error.The @OriginalWorks BETA V2 bot has upvoted(0.5%) and checked this post! https://en.wikipedia.org/wiki/CRISPR
Peace, Abundance, and Liberty Network (PALnet) Discord Channel. It's a completely public and open space to all members of the Steemit community who voluntarily choose to be there.Congratulations! This post has been upvoted from the communal account, @minnowsupport, by Sri Harsha from the Minnow Support Project. It's a witness project run by aggroed, ausbitbank, teamsteem, theprophet0, someguy123, neoxian, followbtcnews/crimsonclad, and netuoso. The goal is to help Steemit grow by supporting Minnows and creating a social network. Please find us in the
If you would like to delegate to the Minnow Support Project you can do so by clicking on the following links: 50SP, 100SP, 250SP, 500SP, 1000SP, 5000SP. Be sure to leave at least 50SP undelegated on your account.