Hi fellow Steemians! Today I am going to teach you a bit more about optogenetics. Quick recap – optogenetics represent new method of controlling cells with high precision in space and time. In order to do so special light-sensitive proteins (termed opsins) have to be expressed in these cells (mainly neurons). Expression means that there is a DNA in the neurons that encodes these specific proteins, so they can be produced. Today I will show you the way gene expression is regulated and how to insert these parts of foreign DNA into neurons.
I already covered some topics in optogenetics, so if you want to catch up, see the links at the end of the article.

Gene expression
One of the advantages of optogenetics is its spatial specificity. This specificity is due to the usage of cell specific promoters. Just to clarify, promoters are regions of DNA that initiate transcription (meaning expression) of certain gene. The expression of opsin can be either directly under the control of promoter (which is active only in certain cells) or binary system might be used. Binary systems are composed from two parts – driver and reporter genes. The expression of driver genes is directly under the control of cell specific promoters. Reporter genes are active only in cells in which are driver genes expressed. An example of a binary system is the usage of Cre recombinase. It catalyses the recombination between two loxP sites (specific homological sequences of DNA). What is recombination you ask? Well, good question and I probably should write article about that as well. But basically, recombination means that two regions of DNA exchange their parts- like, you have two strands of DNA, they recombine and then they are mixture of one-another. Recombination causes either deletion or inversion of DNA localized between these two loxP sites (it all depends on the orientation of loxP sites). The gene coding Cre recombinase is the driver gene in this case. The reporter gene is bounded between two lox sites; it is either an inhibitor of transcription (meaning that it prevents the expression of the gene) or inverted opsin-coding (just to make you remember opsins = light-sensitive proteins) gene. The presence of both Cre recombinase and loxP genetic constructs in one cell causes either dysfunction of transcription inhibitor or inversion of opsin-coding gene to its functional position and so the expression of opsins may begin. I know this is a lot and also quite hard to understand. That is the reason why I created this schemes so you can get better idea what the hell I am talking about.. :D
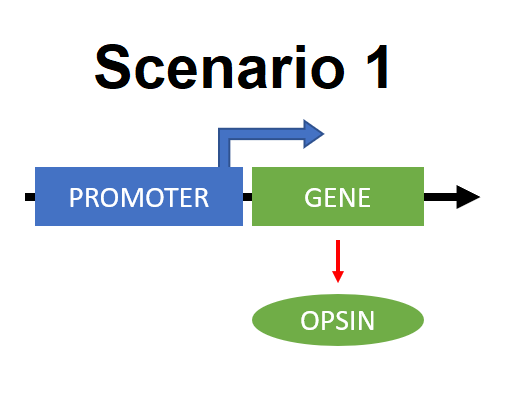
Scenario 1: Gene for light-sensitive protein is directly under the control of promoter.
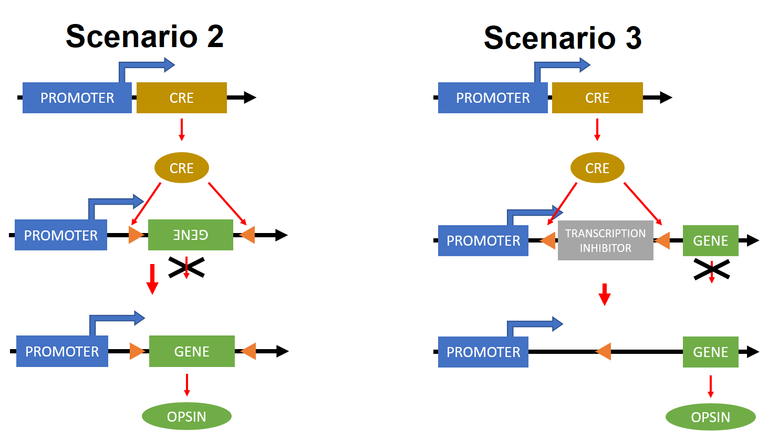
Scenario 2: We have binary system here. The gene coding Cre recombinase is expressed only in specific cells – Cre is termed driver gene in this case. Our light sensitive protein is not expressed, because there is transcriptional inhibitor. Luckily for us there are two loxP sites (orange triangles) around this inhibitor. When Cre recombinase is expressed in cells where is also this transcription inhibitor several things happen. The inhibitor is cut out of the DNA region and the transcription may happen and thus opsin is being expressed.
Scenario 3:LoxP sites are arranged in different fashion here. And our gene of interest is inverted. When Cre recombinase is presented, then this gene gets into correct position and opsin is being expressed. Simple! :D
Gene delivery
There are generally three ways of transportation of genetic constructs to cells of interest. The first is the usage of viral vectors. The expression of proteins is fast and robust. The downside of this strategy is the limited amount of DNA that can be put into a virus. Transgenic animals represent a second option of vectors. Using this approach allows the insertion of larger DNA sequences. The main limitations include the time-consumption of this method and limitation of usable organisms. The third and probably the most effective strategy combines the previous two. It is tightly bound to binary systems. Driver gene coding Cre recombinase has to be expressed only in specific locations of transgenic organism. The virus will carry the reporter gene - the inverted opsin sequence, bounded by two loxP sites. Even though the virus will infect all cells, opsins will be expressed only in cells with driver genes. This strategy combines the high spatial specificity of transgenic approach and the option to use several different opsins in one organism, thus bypassing the time-consuming process of creating a new transgenic line. This might also be kinda hard to understand so again I will try to show you this last strategy on schematics bellow.
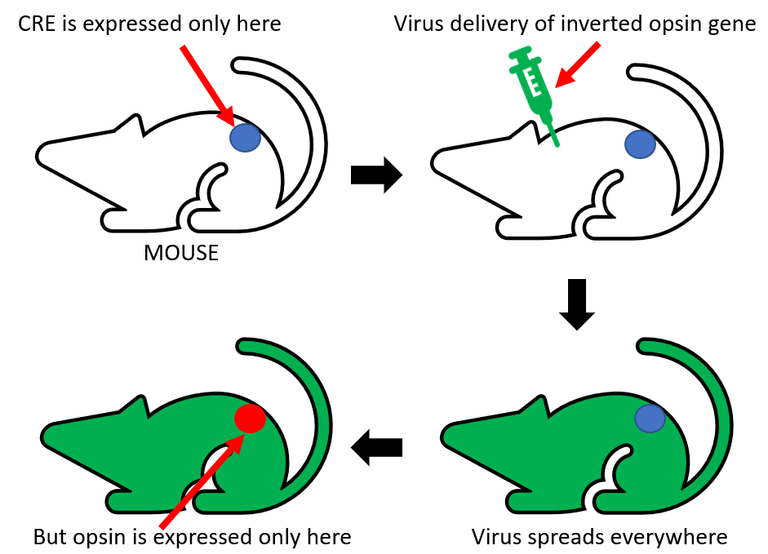
Sooo, I believe you all did learn something new today and hopefully will join me next time as well. Next article from my optogenetics series will introduce light delivery capabilities in optogenetics – fibres and LEDs and stuff. Stay tuned!
Bye! :)
Previous parts of Optogenetics series:
Part 1 – Introduction
Part 2 – History of Optogenetics
Part 3 – Opsins – light sensitive proteins
After 5 years of studying molecular biology and 5-10 years after, it would be smart to study it again...
Crazy fast development. Thank you for this post (and previous one). And several other posts.
Also, you explained very well and clear so the people with no background knowledge can understand as well. Great writing style!
Thank you very much for such positive feedback! I am glad that someone enjoys reading these articles. :)
Well, yes, the progress in science is so rapid that it is impossible to keep track of every technique or new discovery and I believe that this process will be even faster in the future. So, the only option is to really choose something and try to be expert in that particular area, haha.
You know you won't be expert, right?
I love you though.