
(1)
One of my favorite topics to cover on Steemit are new high-tech devices that can revolutionize the scientific community. However, many of these devices won’t have an immediate impact on most people reading these articles because they’re specialized and expensive. Today, I’ll be covering a type of device that’s revolutionary because it’s just the opposite; common and cheap. They’re known as paper-based analytical devices.
What are they
 A litmus test (3).
A litmus test (3).
On second thought, litmus tests aren’t the best example. They’re not too widely used in the lab anymore and you’d only be familiar with them if you’ve taken enough chemistry courses to have been shown them at some point. Here’s a better example:
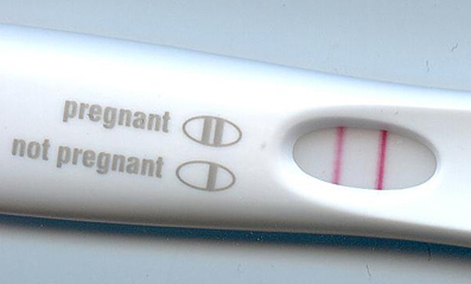
No other two lines have caused as much excitement or dread (4).
Ah the pregnancy test. One of film’s laziest shorthand devices for showing that someone’s been knocked up. That little window showing either one or two lines is actually a strip of paper coated with an antibody designed to detect the pregnancy hormone human chorionic gonadotrophin (5).
Why is this important
Paper-based analytical devices are important because they’re expected to be a core component of the “Lab on a Chip” pipedream that all biotech companies would love to develop. I’ve described the “lab on a chip” concept in an earlier post, so I’ll just quote myself to re-explain.
The idea behind a “lab on a chip” is to be able to perform complex laboratory analysis using a simple instrument in a non-laboratory setting. If such an instrument became a reality we can make major advances in health care and research. With it, a clinic in a third world country can screen for a unique virus in patients in seconds, researchers can obtain real time DNA sequences while sampling environmental sites, and deadbeat dads can get a paternity test without having to go on Murray.
Paper is inexpensive, easy to make, and stable at room temperature. It readily adsorbs liquids and its inert to most compounds and enzymes. The pregnancy test mentioned above is one of the great success stories of these assays. It’s a reliable and inexpensive way for a woman to determine if she’s pregnant within minutes at home. They’re so popular that you can buy them in any pharmacy and see commercials for them on TV.
The only real limitation when developing a paper-based assay is the enzymes and chemicals used for detection. They have to be stable enough to stay active at room temperature and be able to induce a measurable change on the paper when exposed to the reagent it’s designed to detect. This has been the primary challenge facing researchers developing these devices, but progress is being made. Recent articles have demonstrated working paper assays designed to detect tuberculosis (6), identify blood type (7), and assess water quality (8). Below, I’m going to focus on one example that shows just what the development of these assays looks like.
Detecting antibiotic resistance with paper
I’ve chosen to focus on the paper “Utilizing Paper-Based Devices for Antimicrobial Resistant Bacteria Detection” by Katherine E. Boehle et al. because I’ll jump at any opportunity to study bacteria (9). In this paper, the authors wanted to create a paper assay for detecting bacteria in a water sample that could resist beta-lactamase antibiotics. This class of antibiotics includes penicillin, one of the most well know and ubiquitous antibiotics (10). The wide use of these antibiotics has led to a proliferation of resistances and determining whether a pathogen will respond to these antibiotics can be critical information for treating patients.
The key technology behind this assay is a specialized compound called nitrocefin. This compound is normally yellow and contains a beta-lactam ring, a distinct feature of all beta-lactam antibiotics. The useful feature of this compound is that it will turn bright red if the ring is hydrolyzed. Hydrolysis can only occur if the compound comes into contact with bacteria that produce beta-lactamase, an enzyme that confers resistance to these antibiotics.
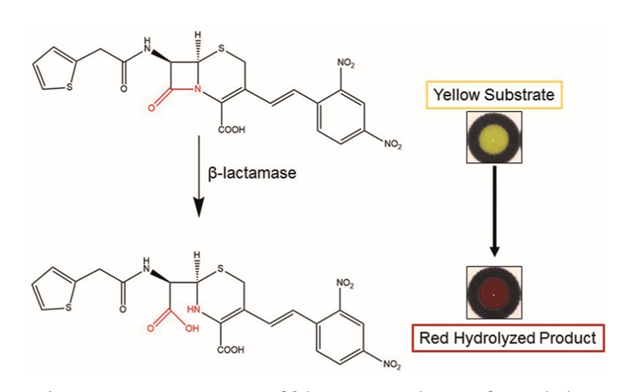
Hydrolysis of nitrocefin by beta-lactamase turns the chemical from yellow to red (9).
The authors act like nitrocefin is just the natural choice for this study, but I’m sure they did extensive testing with a bunch of promising candidate detectors before settling on this one. In fact, if you read between the lines of most papers, you can actually see a graduate student’s life withering away.
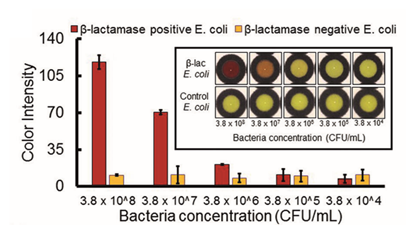 Nitrocefin only turns red if E. coli can express beta-lactamase (9)
Nitrocefin only turns red if E. coli can express beta-lactamase (9)
A color change was only observed when sufficient beta-lactam producing E. coli were added to the paper, thus confirming that nitrocefin was an ideal compound for detecting bacteria with this resistance mechanism.
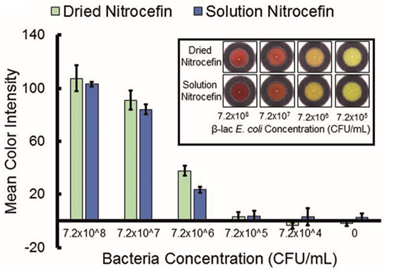 Dried nitrocefin is just as effective as aqueous nitrocefin (9).
Dried nitrocefin is just as effective as aqueous nitrocefin (9).
I’m sure the authors raised a mighty cheer upon getting this result, but their work wasn’t done. They had to bolster this result with supporting data further confirming the usefulness of nitrocefin for a paper based assay. The middle portion of the paper is mostly comprised of these types of tests. For example, they compared to color changing activity of dried nitrocefin to dissolved nitrocefin to confirm that drying the compound didn’t affect the assay (it didn’t).
They also wanted to show that this assay would work in real world sample, so they tested water going into (influent) and out of (effluent) the Drake Water Reclamation Facility located in Fort Collins, Colorado, United States. The idea here is that the sewage water going into the facility will be full of bacteria, some of which are expected to be beta-lactamase resistant, and the treatment process will significantly reduce the number of bacteria in the sample. Detection was possible, and signal intensity was predictably higher in the influent than in the effluent.
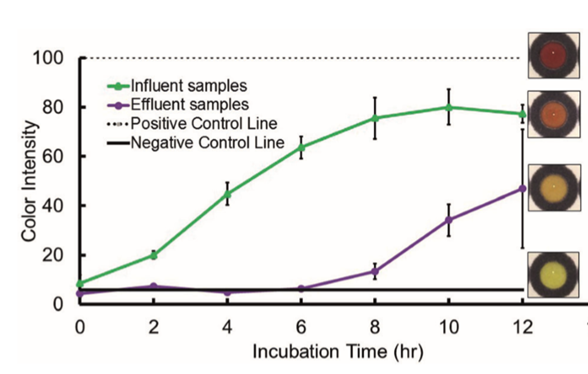
Beta-lactamase activity was detected in water samples going into and out of a waste treatment plant (9).
Finally, they tested individual bacterial species isolated from river water. They wanted to confirm that bacteria found in the wild that are resistant to beta-lactams will turn nitrocefin red. For the most part this experiment worked, one bacteria that was identified as not resistant did not turn nitrocefin red and 6 of 7 bacteria identified as resistant turned nitrocefin red.
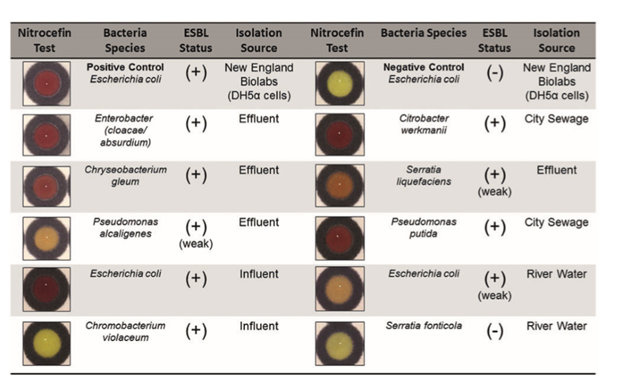
Environmental bacteria known to express beta-lactamase turned nitrocefin red (9)
However, this test was a bit weak in my opinion. They only confirmed resistance with a specialized agar plate that turns colors in the presence of beta-lactamase producing bacteria. They didn’t directly show these isolates to be resistant to a beta lactamase like penicillin with something like a disk diffusion assay. They also had one false negative. Its acceptable overall, but a bit of a weak note to end on.
Despite the lackluster conclusion, this is a solid paper. It convincingly demonstrates that the color changing compound nitrocefin can be used in a paper-based assay to quickly detect beta-lactamase resistant bacteria. Its much faster than conventional methods which require growing the bacteria on a specialized plate or culture media. Its also poised to be much less expensive if mass produced. This probably won’t be put into use immediately, as slightly more rigorous testing is needed to confirm its effectiveness. However, I wouldn’t be too surprised if it becomes a routine part of water quality tests and possibly even pathogen detection in patients soon.
References:
(1) https://www.nature.com/articles/srep08719
(2) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5529145/
(3) https://es.wikipedia.org/wiki/%C3%81cido
(4) https://commons.wikimedia.org/wiki/Pregnancy_test
(5) https://www.ncbi.nlm.nih.gov/pubmed/1639991
(6) https://pubs.acs.org/doi/abs/10.1021/acssensors.7b00450
(7) http://stm.sciencemag.org/content/9/381/eaaf9209.full
(8) https://www.sciencedirect.com/science/article/pii/S0039914017309013
(9) https://onlinelibrary.wiley.com/doi/abs/10.1002/anie.201702776
(10) https://en.wikipedia.org/wiki/B-lactam_antibiotic
Images
All images were taken either directly from the publication referenced or labeled for reuse on Google Images. If any image owner has an issue with this article, please contact me and I will address the issue.

Thank you for your contribution. Dont forget to link references and sources when applicable!
=======================================================================================
This post was upvoted by Steemgridcoin with the aim of promoting discussions surrounding Gridcoin and science.
This service is free. You can learn more on how to help here.
Have a nice day. :)
😄 👍
That's a solid idea.
But, to be implemented, is 6 out of 7 enough? Or is this just a preliminary "proof of concept" type of deal, and now the goal is to improve the accuracy?
I think they need a lot more validation/improvement for this to be implemented. Still, 6 out of 7 is pretty damn good.
Thank for the interesting post. This is certainly a great concept but will need a lots of work to be applicable for clinical settings. Detection limit of 10^6 CFU/ml is very high given that sepsis patients might have 5-100 CFU/ml bacterial load in their blood.
Using paper to detect bacterial... This is a huge step towards the realization of lab on a chip concept. Great work @tking77798 how would i have known tthis
Thanks for the compliment!
This is great technology for sure. Am glad to be part of the strength science community. Am going to learn a lot in the science world and help my country be a better place. Am interested in research and I think this is a great platform for me to catch up. Cheers to technology
Awesome, thanks for reading!
Anything that has antibiotic resistant bacteria has my interest. It's great we are progressing on simpler and easier detection methods.
I also liked your assessment of their work.
Thanks for a beautiful write up !
Thanks man! Glad to have the support!
I didnt know that there is such technology to actually determine the presence of beta lactamase producting organisms apart from the standard culturing. Blood culture results usually cost days to be available and detecting this organism much earlier may help us to change to appropriate antibiotics, which will result in better outcomes for patients. Nice info !
Yeah, earlier detection is one of the major reasons why this is being developed. Hopefully it gets to the market soon.
Besides earlier detection, the lack of refrigeration, liquid handling, multiple reagents, etc make these ideal for developing world testing, which is where they would be most useful.
Absolutely!
Did lots of chemistry in my secondary school days(though hated it but enjoyed the practical more) an the litmus paper was one of my fascinating materials then, you gave in your article a glimpse of how they work and it's a learn for me today.
This paper joining the fight against bacteria will be an accomplishment for our world even in getting bacteria free water, i really did enjoy the piece, thanks for sharing.
Glad you learned something.
A little bit crude, but as the idea, it looks interesting and "refreshing".
It's nice to know that old tricks can find the application even in 2018.
Yeah, its not too elegant at this stage. It might never be, but as long as its cheap and effective it should be put into use.
guau this technology should be share and distributed around the world specially in poor countries, to development a better healthy in the populations in the world to identify bacteries that should kill people
thank you so much for sharing.
Excellent post, right up my alley in water quality analysis techniques.
Glad you like it!
Cool stuff, and hopefully implemented soon.
rofl! sad but so true...
Glad someone caught the joke :D
Congratulations @tking77798! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on any badge to view your own Board of Honor on SteemitBoard.
To support your work, I also upvoted your post!
For more information about SteemitBoard, click here
If you no longer want to receive notifications, reply to this comment with the word
STOPDo not miss the last announcement from @steemitboard!
Great job on this post very interesting
Thanks!
Anytime i gave you a follow
Using paper to detect bacterial... This is a huge step towards the realization of lab on a chip concept. Great work @tking77798 how would i have known tthis
THIS IS GREAT! A salute for you, @tking77798