Repository
https://github.com/BFuks/mad5-utopian-exercises
The files described in this post are saved as:
ex1c-irelandscape/test_cms.cpp
ex1c-irelandscape/saf_reader.py
ex1c-irelandscape/pldoc.py
ex1c-irelandscape/ex1c.py
ex1c-irelandscape/xml_as_dict/
Introduction
third exercise in his series about open-source implementation of LHC particle analysis.This post describes my solution for @lemouth
[Image credits - Pixabay]
In this particular exercise @lemouth introduced us to a new mechanism of particle isolation.
This new isolation procedure is applied to photons in the exercise and consists in calculating the sum of all transverse momenta of charged hadrons (Iπ), neutral hadrons (Iη) and photons (Iγ) within an angular distance dR of the subject photon.
These sums are compared to the transverse momentum of the subject photon and if these are small enough the photon is tagged as a signal photon.
The exercise consisted in isolating signal jets (as explained in the last exercise) and signal photons (as explained above).
The resulting jets and photons transverse momenta and pseudo rapidities are then to be plotted in histogram diagrams.
The Madanalysis5 framework is used to compute the histogram bins and a the charts were to be rendered using Python and Matplotlib.
Exercise Implementation
I will now describe my solution for the exercise.
I'm not entirely sure that I have applied all the right isolation criteria from the research papers so the following definitely needs to be checked.
Signal Particle Isolation and Histograms using MadAnalysis5
The following describes the C++ code implemented.
A new Madanalysis5 project test_cms was created for this purpose.
First, in the Initialize method, I created a "dummy" region selection and added 4 histograms for the photons/jets transverse momenta and pseudo rapidities with 20 bins each.
The range for the momenta is [0, 1000] (GeV).
The range for the photons pseudo rapidity η is [-1.5, 1.5] and [-5, 5] for the jets.
bool test_cms::Initialize(const MA5::Configuration& cfg, const std::map<std::string,std::string>& parameters)
{
cout << "BEGIN Initialization" << endl;
Manager()->AddRegionSelection("dummy");
Manager()->AddHisto("pt_photons", 20, 0, 1000);
Manager()->AddHisto("pt_jets", 20, 0, 1000);
Manager()->AddHisto("n_photons", 20, -1.5, 1.5);
Manager()->AddHisto("n_jets", 20, -5, 5);
cout << "END Initialization" << endl;
return true;
}
Now, in the Execute method, I first apply the weight of the processed event as all events are not necessarily equal:
Manager()->InitializeForNewEvent(event.mc()->weight());
Now I need to isolate jets from electrons. Madanalysis5 provides a JetCleaning method for this but it appears like it required a vector of jets and lepton pointers, instead of vector of objects.
For this reason, I first created these vectors of pointers:
// JetCleaning expects vectors of pointer
for (auto &jet : event.rec()->jets())
jets.push_back(&jet);
for (auto &electron : event.rec()->electrons())
electrons.push_back(&electron);
Then I invoke the JetCleaning method:
// Remove jets electrons overlap
auto cleaned_jets = PHYSICS->Isol->JetCleaning(jets, electrons, 0.2);
I now needed to build a vector of jets meeting the requirement from the research paper. From what I understood such jets need pT > 30GeV and |η| < 5:
In the process, I add each signal jet momentum and pseudo rapidity to the correct histogram.
// Extract jets (pt > 30GeV, |n| < 5)
std::vector<RecJetFormat> signal_jets;
for (auto &jet : cleaned_jets)
{
if (jet->pt() > 30 && jet->abseta() < 5)
{
Manager()->FillHisto("pt_jets", jet->pt());
Manager()->FillHisto("n_jets", jet->eta());
signal_jets.push_back(*jet);
cout << "Adding jet - Pt = " << jet->pt() << ", |n| = " << jet->eta() << endl;
}
}
Now it is time to extract the signal photons.
This involves applying the sum of momenta isolation mechanism mentioned above with the media barrel criteria from table 3 of the photon reconstruction research paper.
Furthermore, each photon needed to satisfy additional requirements with |η| < 1.44 and H/E < 0.05 (the ratio of hadronic to EM energy deposition).
As for jets, the resulting photons momenta and pseudo rapidities are added to the histograms:
// Extract photons (H/E < 0.05, |n| < 1.44 and medium barrel isolation critera)
for (auto &photon : event.rec()->photons())
{
double iPi = PHYSICS->Isol->eflow->sumIsolation(photon,
event.rec(),
0.3,
0.,
IsolationEFlow::TRACK_COMPONENT);
double In = PHYSICS->Isol->eflow->sumIsolation(photon,
event.rec(),
0.3,
0.,
IsolationEFlow::NEUTRAL_COMPONENT);
double Igam = PHYSICS->Isol->eflow->sumIsolation(photon,
event.rec(),
0.3,
0.,
IsolationEFlow::PHOTON_COMPONENT);
auto photon_pt = photon.pt();
if (Igam >= (0.7 + (0.005 * photon_pt)) &&
In >= (1.0 + (0.04 * photon_pt)) &&
iPi >= 1.5 &&
photon.HEoverEE() < 0.05 &&
photon.abseta() < 1.44)
{
cout << "Adding photon: Pt = " << photon_pt << ", |n| = " << photon.eta() << endl;
Manager()->FillHisto("pt_photons", photon_pt);
Manager()->FillHisto("n_photons", photon.eta());
}
}
Building and executing the program results in the following output and a histos.saf file:
=> sample format: Delphes-ROOT file produced by Delphes + MA5tuned-cards.
=> progress: [===> ]
Adding jet - Pt = 215.34, |n| = -0.0193725
Adding jet - Pt = 93.921, |n| = -0.705013
Adding jet - Pt = 45.8521, |n| = -0.0411652
Adding jet - Pt = 35.5923, |n| = -0.0116608
Adding jet - Pt = 33.5812, |n| = -1.90628
Adding jet - Pt = 31.3496, |n| = -2.5429
Adding photon: Pt = 23.8187, |n| = -0.0402587
=> progress: [======> ]
Adding jet - Pt = 182.809, |n| = -0.7438
Adding jet - Pt = 93.1466, |n| = -0.474776
Adding jet - Pt = 90.2955, |n| = -0.818397
Adding jet - Pt = 52.5757, |n| = -2.36295
Adding jet - Pt = 44.9884, |n| = -0.240601
=> progress: [==========> ]
Adding jet - Pt = 221.713, |n| = 1.99956
Adding jet - Pt = 202.161, |n| = 1.76592
Adding jet - Pt = 200.012, |n| = 0.512909
Adding jet - Pt = 60.5474, |n| = 0.883823
Adding jet - Pt = 56.4556, |n| = 2.2267
Adding jet - Pt = 39.5496, |n| = 0.233984
Adding jet - Pt = 37.2858, |n| = 4.63225
Adding jet - Pt = 37.2195, |n| = 1.84628
Adding jet - Pt = 34.2171, |n| = -1.52781
=> progress: [=============> ]
Adding jet - Pt = 112.661, |n| = 0.77616
Adding jet - Pt = 97.7057, |n| = 1.42781
Adding jet - Pt = 69.5077, |n| = 2.37874
Adding jet - Pt = 64.4589, |n| = 3.39025
Adding jet - Pt = 62.6108, |n| = 1.65248
Adding jet - Pt = 53.4359, |n| = 1.30738
Adding jet - Pt = 42.9801, |n| = 0.22429
Adding jet - Pt = 40.8307, |n| = 1.77895
=> progress: [=================> ]
Adding jet - Pt = 257.912, |n| = -0.122587
Adding jet - Pt = 153.493, |n| = 2.22588
Adding jet - Pt = 85.1586, |n| = -0.13745
Adding jet - Pt = 75.27, |n| = 0.116588
Adding jet - Pt = 67.0508, |n| = -2.81922
Adding jet - Pt = 58.5218, |n| = 0.158928
Adding jet - Pt = 50.6759, |n| = -0.267294
Adding jet - Pt = 50.0392, |n| = 0.296783
Adding photon: Pt = 57.1496, |n| = 0.0936919
Adding photon: Pt = 11.9002, |n| = 0.167734
=> progress: [====================> ]
Adding jet - Pt = 282.489, |n| = -0.0644463
Adding jet - Pt = 146.485, |n| = -1.08005
Adding jet - Pt = 115.989, |n| = -1.43669
Adding jet - Pt = 100.48, |n| = -0.807133
Adding jet - Pt = 63.657, |n| = -0.085302
Adding jet - Pt = 49.2006, |n| = 0.505378
Adding photon: Pt = 23.0164, |n| = -0.112591
=> progress: [========================> ]
Adding jet - Pt = 88.2667, |n| = -0.527979
Adding jet - Pt = 82.7576, |n| = -1.44815
Adding jet - Pt = 70.9671, |n| = -0.983675
Adding jet - Pt = 40.0826, |n| = -1.66528
=> progress: [===========================> ]
Adding jet - Pt = 127.115, |n| = -0.415383
Adding jet - Pt = 124.569, |n| = 0.00864341
Adding jet - Pt = 69.6743, |n| = -0.103116
Adding jet - Pt = 58.3917, |n| = -1.85804
Adding jet - Pt = 50.7217, |n| = -1.19539
=> progress: [===============================> ]
Adding jet - Pt = 103.738, |n| = -0.726875
Adding jet - Pt = 64.2951, |n| = -1.91489
Adding jet - Pt = 51.9058, |n| = -1.07069
Adding jet - Pt = 44.0021, |n| = -0.0573528
Adding jet - Pt = 41.2539, |n| = -0.250298
Adding jet - Pt = 36.7969, |n| = -1.39416
Adding jet - Pt = 35.0672, |n| = 1.88652
Adding jet - Pt = 33.101, |n| = -2.63671
Adding photon: Pt = 10.9285, |n| = -0.648636
=> progress: [==================================>]
Adding jet - Pt = 252.772, |n| = 0.455895
Adding jet - Pt = 152.672, |n| = 0.461934
Adding jet - Pt = 68.6698, |n| = 0.292493
Adding jet - Pt = 45.4375, |n| = -0.581494
Adding jet - Pt = 43.5252, |n| = 2.04942
Adding jet - Pt = 33.9009, |n| = 1.41886
Adding photon: Pt = 58.532, |n| = 0.484886
Adding photon: Pt = 24.7049, |n| = 0.466582
=> progress: [===================================]
=> total number of events: 10 ( analyzed: 10 ; skipped: 0 )
* Finalizing all components ...
* Total number of processed events: 10.
BEGIN Finalization
END Finalization
+----------------------------------------------------------------------------------------------------------------------+
| LogReport-Warning |
+----------------------------------------------------------------------------------------------------------------------+
| Message NIterations @ File Line |
|----------------------------------------------------------------------------------------------------------------------|
| the input file has been produced with ROOT v 1 root/ROOTReader.cpp 70 |
+----------------------------------------------------------------------------------------------------------------------+
* Goodbye.
Processing SAF file and generating histograms using Matplotlib
First I installed matplotlib on Ubuntu:
# apt-get install python-matplotlib
The SAF file contains all histograms information in XML format.
I wanted to develop a class aimed at extracting the information in a way that would make it convenient to use this information from a Python program.
I decided to convert all histogram XML sections into Python dictionaries.
To do this I used Duncan McGreggor's Python recipe which I stored under a xml_as_dict folder.
I then proceeded in implementing two sets of python classes in distinct .py files: saf_reader.py and pldoc.py.
saf_reader.py implements a class SafReader that can be used to read and process the XML document in the SAF file and produce a list of SafHisto instances, each one storing information about a single histogram. The SafHisto class provides convenient methods to return the title, range and bins of the histogram.
pldoc.py implements a PlFigure class which provides a convenient way to generate a configurable number of charts in a Matplotlib figure arranged in a grid like manner. When instantiating the class, the user code specifies how many rows and columns of charts the figure should have.
PlFigure provides an AddBarChart method which takes, among other things, a SafHisto instance as argument.
Note that PlFigure does not refer directly to saf_reader.py so any histogram class providing the Bins, Title and Range method will work. This might be convenient in the future to generate histogram plots from non-SAF sources.
Here is the code for saf_reader.py
import xml_as_dict.xml_as_dict as xml_as_dict
import xml.etree.ElementTree as ET
# A class to hold information about a single Histogram stored
# in SAF file.
class SafHisto :
# histo is a dictionary conversion of the SAF XML
def __init__ (self, histo) :
self.histo = histo
def Title (self) :
description = self.histo['Description']
# Parse the description to retrieve title
elems = description.split('"')
if len(elems) > 1 :
return elems[1]
else :
return ''
# Return xmin, xmax
def Range (self) :
description = self.histo['Description']
# Parse the description to retrieve xmin and xmax
pos = description.find('nbins')
pos += description[pos:].find('\n') + 1
fields = description[pos:].split()
return float(fields[1]), float(fields[2])
# Return all bin data
def Bins (self) :
underflow = 0.0
overflow = 0.0
data = self.histo['Data']
bins = []
for row in data.split('\n') :
if len(row.strip()) == 0 :
continue
cols = [ float(x) for x in row.split()[:2] ]
bins.append(cols[0] - cols[1])
return bins
class SafReader () :
def __init__ (self, file) :
with open(file) as f :
# Store all histograms in a list
self.histos = [ SafHisto(x) for x in self._GetHistos(f.read()) ]
f.close()
# Extract all histograms from the SAF XML
# Each histogram is stored as a Python dictionary corresponding to
# The <Histo></Histo> section of the XML.
def _GetHistos (self, string) :
histos = []
start = 0
while True :
start = string.find('<Histo>', start)
if start == -1 :
break
end = string.find('</Histo>', start + 1)
if end == -1 :
break
end += len('</Histo>')
root = ET.fromstring(string[start : end])
histos.append(xml_as_dict.XmlDictConfig(root))
start = end + 1
return histos
def GetHistos (self) :
return self.histos
And the pldoc.py file:
import numpy
import matplotlib.pyplot as plt
from matplotlib import colors
from matplotlib.artist import setp
from matplotlib.backends.backend_pdf import PdfPages
class PlFigure (object) :
def __init__ (self,
nrows = 1,
ncols = 1,
figsize = (5, 6),
backend = 'svg') :
plt.switch_backend(backend)
self.ncols = ncols
self.current_plot = 0
# Create a grid of plots on the figure
self.fig, self.subplots = plt.subplots(nrows = nrows,
ncols = ncols,
figsize = figsize,
dpi=240)
# Adjust spacing between plots
self.fig.subplots_adjust(wspace = 0.4, hspace = 0.4)
i = 1
for row in range(nrows) :
for column in range(ncols) :
# Add some formatting of the axis
self.subplots[column][row].ticklabel_format(style = 'sci',
scilimits = (0.01,100),
axis = 'both')
++i
# Get the next unused plot
def _GetNewPlot (self) :
column = self.current_plot % self.ncols
row = int(self.current_plot / self.ncols)
self.current_plot += 1
subplot = self.subplots[column][row]
return subplot
# Create a bar chart from histogram data
def AddBarChart (self,
histogram,
xaxis_label = None) :
subplot = self._GetNewPlot()
bins = histogram.Bins()
title = histogram.Title()
min, max = histogram.Range()
# Calculate the x values and the width of each bar
x = numpy.linspace(min, max, len(bins))
width = (max - min) / len(bins)
# Draw the chart
subplot.bar(x, bins, width)
subplot.set_title(histogram.Title())
subplot.grid(color='grey', linestyle='-', linewidth=0.5)
subplot.set_axisbelow(True)
if xaxis_label :
subplot.set_xlabel(xaxis_label)
def show (self) :
self.fig.show()
def save (self, filepath, **kwargs) :
self.fig.savefig(filepath, **kwargs)
As you can see above the application can save the resulting charts in a file using the PlFigure.save method. The supplied file name can indicate any of the formats supported by Matplotlib, including svg, png, jpg, etc.
Finally here is the main application code (implemented in ex1c.py:
import sys
from saf_reader import SafReader, SafHisto
import pldoc
reader = SafReader(sys.argv[1])
i = 0
xaxis_labels = ('pT', 'pT', r'$\eta$', r'$\eta$')
fig = pldoc.PlFigure(nrows = 2, ncols = 2)
for histo in reader.GetHistos() :
fig.AddBarChart(histo, xaxis_labels[i])
i += 1
fig.save(sys.argv[2])
The script can then be executed with the following command:
# python ./ex1c.py <saf_file> <output_file>
Result
This result in the following charts:
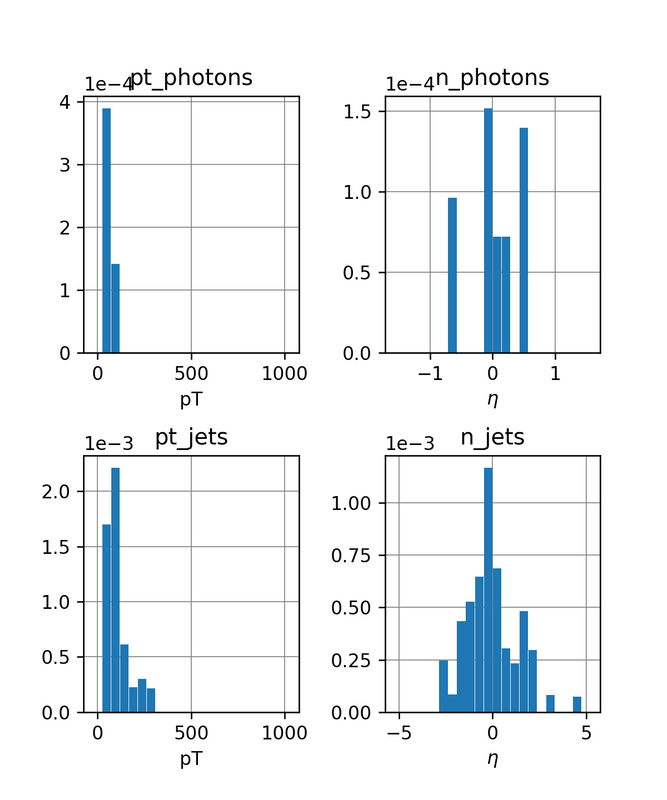
Conclusions
This was a fun exercise though, again, I'm not sure if the final result is correct or not!
I enjoyed learning about Matplotlib and look forward to produce more interesting diagrams in future exercises.
Resources
- # Particle physics @ utopian-io - Objects isolation, histogramming and a first task request by @lemouth
- Search for new physics in the monophoton final state in proton-proton collisions at √ s = 13 TeV
- Performance of photon reconstruction and identification with the CMS detector in proton-proton collisions at √ s = 8 TeV
- Matplotlib.
- XML AS DICTIONARY (PYTHON RECIPE) by Duncan McGreggor
- MadAnalysis5 Download
Hello @irelandscape
Participating in @lemouth's exercise alone is worthy of commendation, how much more writing down all these codes. I am not a student or fan of particles physics but the richness of this article perhaps demonstrates your rich knowledge of the subject matter.
Wishing the very best as you participate in this exercise
Regards
@eurogee of @euronation and @steemstem communities
Thank you as always @eurogee.
Thanks for your nice message!
Thank you for your submission. I would like to guide you to improve the quality of your contributions and inform about expectations of the category.
Even though we welcome contributions to MadAnalysis through exercises @lemouth shares, contributions in the category still should be based on editorial content with good presentation. Considering that blog posts are expected to promote the projects, they need to contain more than code and analysis reports to really add value to the project as a blog post.
Even though I appreciate all the contributions to MadAnalysis project as a science lover, the content should be revised to fit the category.
The first possible improvement would be referring to files in a repository instead of bulk sharing the code or output in the files. Then text to code ratio could be optimized for a good presentation. And beside these, using blogging language for providing an overview would increase the quality without a doubt.
While preparing your future contributions, please do not forget that these post are meant to promote the project in their own ways.
Your contribution has been evaluated according to Utopian policies and guidelines, as well as a predefined set of questions pertaining to the category.
To view those questions and the relevant answers related to your post, click here.
Chat with us on Discord.
[utopian-moderator]Need help? Write a ticket on https://support.utopian.io/.
Hi, thank you for your review.
I note all your points.
I'm still unfamiliar with the format of posts under utopian-io/blog.
I'll have a look at other posts in this category to get a better idea.
I guess the intention behind adding the output of the program to the post was to allow @lemouth to spot quickly mistakes in the implementation. In the future I can commit a file to the repository with the output of the program.
The files added to github are at the beginning of the post.
Cheers.
Congratulations @irelandscape! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on the badge to view your Board of Honor.
If you no longer want to receive notifications, reply to this comment with the word
STOPTo support your work, I also upvoted your post!
Do not miss the last post from @steemitboard!
Participate in the SteemitBoard World Cup Contest!
Collect World Cup badges and win free SBD
Support the Gold Sponsors of the contest: @good-karma and @lukestokes
Amazing..Trying to understand the code. What is it trying achieve or possibly explain?
Good job, keep it coming.
Hi, the code tries to solve @lemouth's exercise which isolate particles for further analysis.
The resulting particles properties are then graphed in histograms.
Thanks!
Thanks for contributing on Utopian.
We’re already looking forward to your next contribution!Hey @irelandscape
Want to chat? Join us on Discord https://discord.gg/h52nFrV.
Vote for Utopian Witness!